Use this graph to display my internet connection stability.

Any large genome-wide association study corrects for population structure, in addition to restricting the sample to subjects of caucasian ethnicity. This is for the most part a practical necessity since mankind’s migration pattern across the globe has left its marks on our genome. Not correcting for these effects would lead to false-positive genetic associations. However, the topic of correcting for these population structure effects has for the most part not reached the field of neuroimaging.
It has been shown in earlier work that head and brain morphology is markedly different between continental genetic ancestries (paper). In a recent work we are showing that one can predict continental ancestry even from resting-state functional MRI. We have not established yet the reason for why this works, but we hypothesize that artifacts from head and brain morphology influence the observed connectivity pattern (paper). This highlights that neuroimaging data is susceptible to differences between genetic ancestries, i.e., population structure.
Do we need adjustment for population structure in neuroimaging? Yes, I think we should be more mindful about these effects and consider including genetic ancestry as covariates in imaging studies. The very same way we are addressing other confounding variables.
We have a PhD studentship available that is focused on uncovering the imaging and genetic basis of drug-resistant epilepsy. More details are available here.
Marzia’s paper with the title “Genetic study of multimodal imaging Alzheimer’s disease progression score implicates novel loci” has been published in Brain. Congrats!
Abstract:
Identifying genetic risk factors underpinning different aspects of Alzheimer’s disease has the potential to provide important insights into pathogenesis. Moving away from simple case-control definitions, there is considerable interest in using quantitative endophenotypes, such as those derived from imaging as outcome measures. Previous genome-wide association studies of imaging-derived biomarkers in sporadic late-onset Alzheimer’s disease focused only on phenotypes derived from single imaging modalities. In contrast, we computed a novel multi-modal neuroimaging phenotype comprising cortical amyloid burden and bilateral hippocampal volume. Both imaging biomarkers were used as input to a disease progression modelling algorithm, which estimates the biomarkers’ long-term evolution curves from population-based longitudinal data. Among other parameters, the algorithm computes the shift in time required to optimally align a subjects’ biomarker trajectories with these population curves. This time shift serves as a disease progression score and it was used as a quantitative trait in a discovery genome-wide association study with n = 944 subjects from the Alzheimer’s Disease Neuroimaging Initiative database diagnosed as Alzheimer’s disease, mild cognitive impairment or healthy at the time of imaging. We identified a genome-wide significant locus implicating LCORL (rs6850306, chromosome 4; P = 1.03 × 10−8). The top variant rs6850306 was found to act as an expression quantitative trait locus for LCORL in brain tissue. The clinical role of rs6850306 in conversion from healthy ageing to mild cognitive impairment or Alzheimer’s disease was further validated in an independent cohort comprising healthy, older subjects from the National Alzheimer’s Coordinating Center database. Specifically, possession of a minor allele at rs6850306 was protective against conversion from mild cognitive impairment to Alzheimer’s disease in the National Alzheimer’s Coordinating Center cohort (hazard ratio = 0.593, 95% confidence interval = 0.387–0.907, n = 911, PBonf = 0.032), in keeping with the negative direction of effect reported in the genome-wide association study (βdisease progression score = −0.07 ± 0.01). The implicated locus is linked to genes with known connections to Alzheimer’s disease pathophysiology and other neurodegenerative diseases. Using multimodal imaging phenotypes in association studies may assist in unveiling the genetic drivers of the onset and progression of complex diseases.
Our paper with the “Genetic study of multimodal imaging Alzheimer’s disease progression score implicates novel loci” was accepted for publication in Brain!
Authors: Marzia A. Scelsi, Raiyan R. Khan, Marco Lorenzi, Leigh Christopher , Michael D. Greicius, Jonathan M. Schott, Sebastien Ourselin, Andre Altmann
A Related abstract at OHBM 2017 can be found here.
 Our paper with the title “Predicting outcome in idiopathic pulmonary fibrosis using automated CT analysis” was accepted in AJRCCM!
Our paper with the title “Predicting outcome in idiopathic pulmonary fibrosis using automated CT analysis” was accepted in AJRCCM!
Authors:
Joseph Jacob, Brian J. Bartholmai, Srinivasan Rajagopalan, Coline H.M. van Moorsel, Hendrik W. van Es, Frouke T. van Beek, Marjolijn H.L. Struik, Maria Kokosi, Ryoko Egashira, Anne Laure Brun, Arjun Nair, Simon L.F. Walsh, Gary Cross, Joseph Barnett, Angelo de Lauretis, Eoin P. Judge, Sujal, Desai, Ronald Karwoski, Sebastien Ourselin, Elisabetta Renzoni, Toby M. Maher, Andre Altmann, Athol U. Wells
Our abstract “Rare genetic events in sporadic Alzheimer’s disease: a network propagation approach” was selected for an oral presentation at OHBM 2018, Singapore!
On top of that lead author Marzia Scelsi received (again!) a Merit Abstract Award for the 2018 OHBM Annual Meeting. Congrats Marzia!
M. A. Scelsi, J. E. Iglesias, J M Schott, S Ourselin, A Altmann, “Rare genetic events in sporadic Alzheimer’s disease: a network propagation approach”
Introduction
The discovery of rare genetic mutations in the APP, PSEN1, and PSEN2 that cause familial Alzheimer’s disease (AD) has had profound implications for our understanding of deisease pathognesis. There is much interest in investigating rare genetic variants that might increase risk for, or influence the phenotype of, sporadic AD. This however remains challenging due to the very large sample sizes required and the costs of performing whole-genome sequencing (WGS) in these cohorts. Early successes have included the discovery that TREM2 influences risk (Guerreiro et al. 2013), a discovery that has intensified interest in inflammation in AD. Studies such as the Alzheimer’s Disease Sequencing Project (Bis et al. 2017) continue to address this challenge. Here, we present a method to investigate the effects of rare variant in moderately sized sporadic AD cohorts, such as ADNI Our approach is based on signal diffusion on a gene-interaction network, inspired by its successful application to cancer subtyping (Hofree et al. 2013). We sought to develop an integrative approach to guide gene-based association testing with the knowledge of tissue-specific interaction networks.
Our paper with the title “Susceptibility of brain atrophy to TRIB3 in Alzheimer’s disease, evidence from functional prioritization in imaging genetics” is now available online at PNAS!
Read the TIG news story here!
Summary:
In this study, we use an experimental imaging–genetics approach for investigating the genetic underpinnings of brain atrophy in Alzheimer’s disease. We successfully combined state-of-the-art imaging–genetics methods and experimental gene expression data to uncover biology in brain atrophy. The experimental paradigm highlighted a significant role of tribbles pseudokinase 3 (TRIB3) in modulating the typical pattern of Alzheimer’s brain pathology. This result corroborates through rigorous data-driven statistical methods evidence emerging from previous studies about the role of TRIB3 in modulating known mechanisms of neurodegeneration, such as neuronal death, cellular homeostasis, and interaction with established genes causing autosomal dominant Alzheimer’s disease: APP and PSEN1. The developed integrated statistical–experimental methodology could serve as a roadmap for investigations in other disorders.
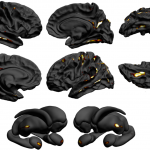
The ENIGMA Epilepsy paper has been published in Brain today!
See also UCL’s press release here.
Whelan et al., 2018: “Structural brain abnormalities in the common epilepsies assessed in a worldwide ENIGMA study”
 Abstract
Abstract
 Marzia was interviewed about her newsworthy OHBM 2017 contribution!! Click here to read the interview!
Marzia was interviewed about her newsworthy OHBM 2017 contribution!! Click here to read the interview!